Integration into relacs¶
 Relacs is a highly configurable recording tool for
electrophysiology.
Relacs is a highly configurable recording tool for
electrophysiology.
It is one use-case showing how to integrate automated data storage and data annotation into the recording process.
Relacs overview¶
In the Neuroethology group relacs is used to record the activity of single neurons during electrophysiological experiments. It controls the experiment, presents stimuli, performs online analyses, adapts stimuli in a closed loop fashion, and stores data to file. Being configurable implies that settings and properties are not forseeable at compile time. Accordingly it requires a lot of flexibility on the side of the storage backend. At the same time it knows a lot of metadata that are essential for corretly analyzing the data.
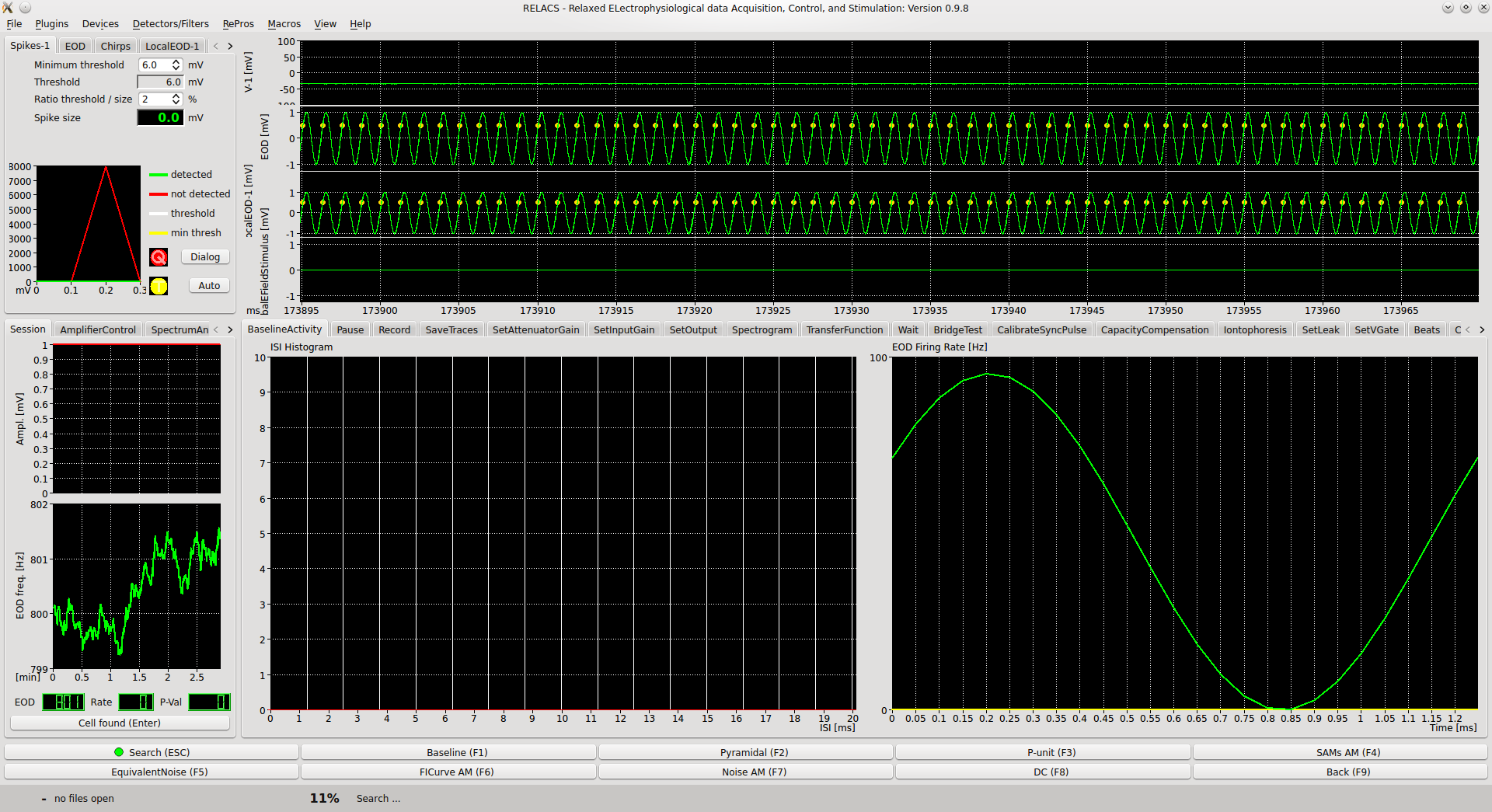
Figure 1: The relacs user interface. The UI is divided into four panels. Top left: Filter and detector plugins that are used to detect events in the recorded data (such as action-potentials, spikes). Top right: Main panel showing the four recorded traces (membrane potential, fish’s electric organ discharge global, same at a reference location, the stimulus applied during the experiment) yellow dots indicate detected events. Bottom left: diverse control panels, or other displays. Bottom right: Space the currently active research protocol (RePro) can use to display data or interact with the experimenter.¶
In the depicted use-case, several traces are recorded in parallel (usually sampled with 20, 40, or 100 kHz per channel 16 bit, depending on experimental requirements).
The neuron’s membrane voltage (V-1).
The fish’s electric organ discharge (EOD), global measurement.
The local measurement of the EOD (LocalEOD-1).
The stimulus put into the tank (GlobalEFieldStimulus).
The times of detected spikes (Spikes-1).
Sometimes the times of EOD discharges and detected chirps (communication signals).
Metadata¶
Relacs knows a lot of metadata that are either configured before the experiment was started (for example the parameters used when measuring a firing rate vs intensity curve, the left figure below), are automatically adapted, or have to be entered by the user when storing the data (right figure below) or are derived from the recorded data (average firing rate, the EOD frequency, etc).
Figure 2 |
Figure 3 |
A recording session¶
Once the recording of a neuron has been established, we are ready to start running the actual experiment. Either pre-defined macros are used to control the experiments or the experimenter starts research protocols (RePros) manually.
During a recording session, the “data time” is not necessarily real time. Data time proceeds only when the acquisition is active and data is actually recorded and stored to disk, this is under control of the currently active research protocol (RePro). Figure 4 below illustrates this.
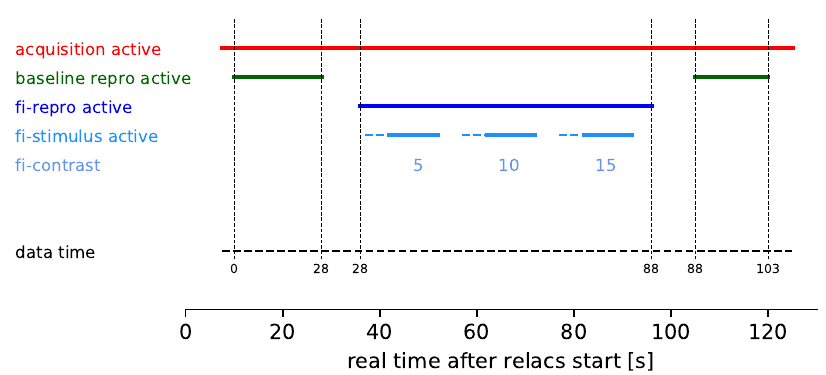
Figure 4: A recording session becomes active once the acquisition is switched on (red bar). Data acquisition, however, data is stored in the NIX file, when the currently running RePros demands it. In this example session there are two RePros active, the so called “Baseline” (green) and the “fi-curve” (blue). Within the time the fi-curve RePro is active three stimuli are presented (light blue). For each stimulus presentation the fi-curve RePro applies a stimulus with a different intensity (contrast, blue numbers).¶
Data storage using NIX¶
So much for the background. In the following we will illustrate how such data is persisted in NIX files and how to work with the data. Since relacs is programmed in C++ the nixio c++ library is used for this.
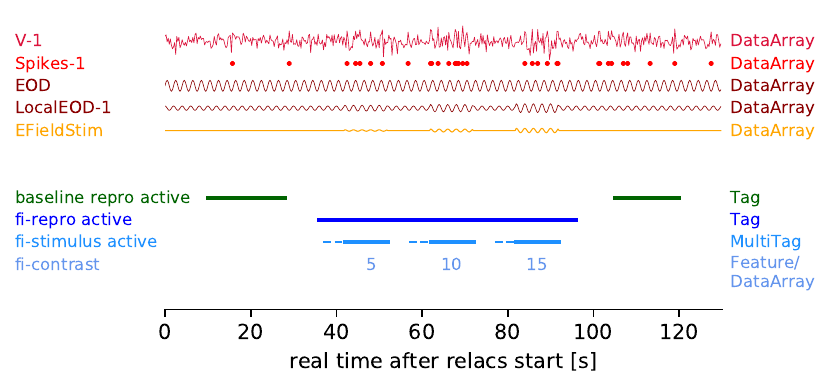
Figure 5: All four recorded traces and all event traces (only
the spike events are shown here) are stored in
nix::DataArrays. Whenever a RePro is active a nix::Tag is
created that stores the start time and temporal extent (in data
time, see above) and links to metadata nix::Section that stores
the static RePro parameters, i.e. those that do not change during
this RePro run. A RePro may apply certain stimuli (the fi-curve
RePro) and these stimulus times and extents are stored in a
nix::MultiTag entity. It further links to some more metadata
entries. Some parameters, in this case the stimulus “contrast”
changes with each stimulus presentation. This information is not
stored in the metadata but is considered a feature of the
stimulus presentation. That is, we create a nix::DataArray to
store these numbers, one for each stimulus presentation, and link
it to the nix::MultiTag as an indexed nix::Feature (see
adding features for details).¶
Storing of continuously sampled data¶
The continuous traces in figure 5 are continuously sampled 1-D
vectors. This only dimension represents time, the measured values
themselves are double values. In NIX data is stored in
nix::DataArray entities. The regular sampling of the time
dimension is defined using a nix::SampledDimension (see storing
data for more information). Since the duration
of the recording is unknown the data is continuously written to file.
The following code block illustrates the basic steps (the full implementation can be found here ):
1void SaveFiles::NixFile::initTraces ( const InList &IL )
2{
3 for ( int k=0; k<IL.size(); k++ ) {
4 NixTrace trace;
5 string data_type = "relacs.data.sampled." + IL[k].ident();
6 trace.data = root_block.createDataArray( IL[k].ident(), data_type, nix::DataType::Float, {4096} );
7 std::string unit = IL[k].unit();
8 nix::util::unitSanitizer( unit );
9 if ( !unit.empty() && nix::util::isSIUnit( unit ) ) {
10 trace.data.unit( unit );
11 } else if ( !unit.empty() ) {
12 std::cerr << "NIX output Warning: Given unit " << unit << " is no valid SI unit, not saving it!" << std::endl;
13 }
14 if ( !IL[k].ident().empty() )
15 trace.data.label( IL[k].ident() );
16 nix::SampledDimension dim;
17 dim = trace.data.appendSampledDimension( IL[k].sampleInterval() );
18 dim.unit( "s" );
19 dim.label( "time" );
20 trace.index = IL[k].size();
21 trace.written = 0;
22 trace.offset = {0};
23 traces.push_back(std::move( trace ));
24 }
25}
In this code snippet IL is a vector of relacs::InData
objects. These provide information about the data coming in.
Line 4: for each k th element of the InputList, a NixTrace
object is created that buffers the entity for later re-use.
Line 6: a nix::DataArray is initialized with a name, a type
(line2), the data type and an initial size. Selecting the initial size
defines the chunksize applied by the underlying HDF5
library. Selecting a too small chunk size will cause performance
problems. Lines 16–19 set the dimension information
(nix::SampledDimension).
At this point, no data has been written to file.
Writing actually happens in a small helper function that works with
the NixTrace object created before:
1void SaveFiles::NixFile::writeChunk( NixTrace &trace, size_t to_read, const void *data )
2{
3 typedef nix::NDSize::value_type value_type;
4 nix::NDSize count = { static_cast<value_type>( to_read ) };
5 nix::NDSize size = trace.offset + count;
6 trace.data.dataExtent( size );
7 trace.data.setData( nix::DataType::Float, data, count, trace.offset );
8 trace.index += to_read;
9 trace.written += to_read;
10 trace.offset = size;
11}
Storing of event data¶
Neuronal events such as action potentials are stored in
nix::DataArrays. Again, the data is basically a 1-D vector in
which the one dimension represents time.
nix::DataArrays for events.¶ 1void SaveFiles::NixFile::initEvents( const EventList &EL, FilterDetectors *FD )
2{
3 for ( int i = 0; i < EL.size(); i++ ) {
4 if ( (EL[i].mode() & SaveTrace) == 0 ) {
5 continue; //Nothing to save
6 }
7 NixEventData ed;
8 ed.el_index = i;
9 ed.index = EL[i].size();
10 ed.offset = {0};
11 std::string ident = EL[i].ident();
12 std::string data_type = "relacs.data.events." + ident;
13 if ( root_block.hasDataArray( ident ) )
14 ident = EL[i].ident() + "_events";
15 ed.data = root_block.createDataArray( ident, data_type, nix::DataType::Double, {256} );
16 ed.data.unit( "s" );
17 ed.data.label( "time" );
18 ed.data.appendAliasRangeDimension();
19 ed.input_trace = FD->eventInputTrace( i );
20 events.push_back(std::move( ed ));
21 }
22}
This is very similar to the initTaces above. Two things are wroth
mentioning. Line 14: The initial size was chosen to be much
smaller, we expect less events than data points in the traces. Line
17: The values stored in the nix::DataArray are times, (in
data time, see above). Thus, we apply an
nix::AliasRangeDimension which indicates that the values
themselves define the dimension (see dimension documentation for details on dimensions).
Noting RePro runs¶
Whenever a RePro is started the start time (data time) is noted and
nix::Tag is created. This indicates when and how long a RePro ran
and links to respective metadata (settings of the RePro). The Tag
refers to all event and continuous data traces (as references
see here for more information of tagging). This allows
for automatic data retrieval of the thus tagged slabs of the data (below).
Defining stimulus segments¶
Within a RePro run relacs may present stimuli. Stimulus epochs are
stored within a nix::MultiTag. Other than the simpler nix::Tag,
the MultiTag is meant to tag multiple regions of interest in the data
that belong together.
If a RePro puts out a number of identical stimuli, start times and
extents are stored in the same MultiTag. A RePro might also apply
stimuli that are not identical due to different parameters. In this
case there will be a nix::MultiTag for each unique stimulus
parametrization.
In relacs RePro options can be given the OutData::Mutable flag,
which communicates that the respective parameter is intended to change
during the RePro run. All mutable parameters are stored as an
nix::LinkType::indexed nix::Feature (see f-i curve example in
figure 5). With this approach, the parameters become directly
available during further data processing (see below).
Working with relacs-flavored NIX files¶
For the following we will use this file containing simulated data. The simulation
models the neuronal activity of a p-type electroreceptor afferent in
the electrosensory system of the weakly electric fish Apteronotus
leptorhynchus. The code snippets shown below are part of
this script which uses the python
library (nixpy). To run the
example code nixpy, numpy, and matplotlib python packages are required.
A simple plot¶
The first example simply plots a short part of the recorded traces.
1def plot_data_snippet(filename, trace_name="V-1", samples=5000):
2 f = nix.File.open(filename, nix.FileMode.ReadOnly)
3 b = f.blocks[0]
4 trace = b.data_arrays[trace_name]
5 dim = trace.dimensions[0]
6
7 plt.plot(dim.axis(samples), trace[:samples], label=trace.name)
8 plt.xlabel("%s [%s]" % (dim.label, dim.unit))
9 plt.ylabel("%s [%s]" % (trace.label, trace.unit))
10 plt.legend()
11 plt.show()
12
13 f.close()
All information that is needed to create a basic plot of the stored
data is attached to the DataArray (trace). Its label and
unit define the y-labels, the dimension descriptor
(nix::SampledDimension) contains all information that is needed to
label the x-axis. In line 7 a convenience function of the dimension
descriptor (see dimensions) is used to
get a vector of time values matching the data.
In this code snippet we use some prior information about the recorded
data (we directly ask for the “V-1” trace). To find out which data
traces are available we can search for nix::DataArrays that have
the type relacs.data.sampled (see defined types below) by using
list comprehension executing:
sampled_traces = [da for da in b.data_arrays if "relacs.data.sampled" in da.type]
for t in sampled_traces:
print(t.name, t.type)
Accessing metadata¶
Relacs knows a lot about the data and settings, this information is
automatically saved to file and is attached to the only nix::Block
entity.
The following code snippte shows how to access metadata about the subject that was used in this recording session.
1def print_subject_metadata(filename):
2 f = nix.File.open(filename, nix.FileMode.ReadOnly)
3 b = f.blocks[0]
4 sections = b.metadata.find_sections(lambda sec : "subject" in sec.type.lower())
5 for sec in sections:
6 sec.pprint(max_depth=-1)
7 f.close()
Two things are worth noting: Line 4 uses the section ‘s find
method to find the “subject” section. The method takes a filter
function as input argument. Here we filter on the section type. Line
6 uses the section’s pprint function to dump an overview to the
command line. The max_depth=-1 argument defines that the full tree
is recursively traversed.
Defined types and their meaning¶
In the nix DataModel almost all entities have a name, type, and an id. Name is a user-specified string, id an is auto-generated UUID and the type is meant to provide semantic meaning. The type support in relacs is not very elaborated which is to some extent due to the flexibility of the tool. For example, relacs on its own, has no information whether an recorded trace is the membrane voltage, a temperature or any other kind of measurement.
The following types are used:
type |
entity |
meaning |
|---|---|---|
relacs.recording |
Block |
Only one
nix::Block is created in each file,all data entities are children of this block and it
represents a “recording” session.
Session names are created from the date and a suffix.
|
relacs.data.sampled |
DataArray |
Regularly sampled data, vectors of time.
nix::SampledDimension as dimension descriptor. |
relacs.data.events |
DataArray |
Any kind of event data, e.g. action potentials, etc.
Entries denote the time at which the event occurred
nix::AliasRangeDimension defines the time dim. |
relacs.stimulus.segment |
MultiTag |
Tags the data segments in which a stimulus was
presented. One position and extent entry for
each segment in which the identical stimulus was
used. This entity is re-used whenever the same stim
is presented, even within a different RePro run.
|
relacs.stimulus.onset |
DataArray |
Onset times of a stimulus segment(s).
nix::SetDimension defines only dimension. |
relacs.stimulus.duration |
DataArray |
Temporal duration of the stimulus segment(s).
nix::SetDimension defines only dimension. |
relacs.repro |
Section |
Metadata containing RePro settings and properties. |
relacs.repro_run |
Tag |
Tags the data segment in which a RePro was active and
links to metadata. This does not imply that a stim-
ulus was active.
|
relacs.repro_group |
Group |
Group that contains all entities created during a
RePro run.
|
relacs.feature.time |
DataArray |
A set of timestamps
nix::SetDimension defines only dimension. |
relacs.feature.amplitude |
DataArray |
A feature of the stimulus segment that is the
amplitude of the stimulus.
nix::SetDimension defines only dimension. |
relacs.feature.mutable |
DataArray |
These features contain the values of settings that
are intended to change. Changes in such settings
will not lead to the creation of a new MultiTag.
nix::SetDimension defines only dimension. |
relacs.feature.repro_tag_id |
DataArray |
Notes the RePro run (the Tag above) during which the
stimulus was active. Contains the Tag’s entity id.
nix::SetDimension defines only dimension. |

